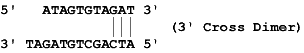All of the formulas used in the program are given below for quick reference:
The rating of a primer provides a quick way of measuring the predicted efficiency of a primer as well as choosing between closely matched primers. The higher the rating of a primer, the higher its amplification efficiency.
The rating of individual primers is calculated as:
Rating = 100 + (DG (Dimer) * 1.8 + DG (Hairpin) * 1.4)
Example: Say a primer has worst DG (Dimer) as -2.4 and worst DG (Hairpin) as -1.0, that primer will have a rating of 94.
The molecular weight of primer is the calculated molecular weight determined using the standard values of molecular weight of individual nucleotides. It is assumed that 5' end of the oligonucleotide is not phosphorylated, which is a common practice. The values used here are:
dG = 329.21 dA = 313.21
dC = 289.19 dT = 304.20
The molecular weight is calculated using the formula:
Molecular Weight of primer = [(Sum of weights of individual nucleotides + 18.02 (for water))-80.00]
Example: Say the primer sequence is ATCGATACGTAG. The molecular weight of this primer will be 4 * (Mol. Wt. of A) + 3 * (Mol. Wt. of T) + 2 * (Mol. Wt. of C) + 3 * (Mol. Wt. of G) + 18.02. = 3749.47- 80.00 = 3669.47
The melting temperature is calculated using the formula based on the nearest neighbor thermodynamic theory. It is the temperature at which half of the oligonucleotides are bonded. The formula is from the paper by Freier et. al. These are the latest and most accurate nearest neighbor based Tm calculations.
Tm = DH/(DS + R * ln(C/4)) + 16.6 log ([K+]/(1 + 0.7 [K+])) - 273.15
DH is enthalpy for helix formation.
DS is entropy for helix formation.
R is molar gas constant (1.987 cal/°C * mol)
C is the nucleic acid concentration.
[K+] is salt concentration.
Example: Say the primer sequence is ATCGATACGTAG. The DH and DS values of this primer will be -85000 cal/mol and -234.7 cal/°K/mol respectively (as calculated below). After substituting all the values, the Tm value of this primer will be 16.69 °K.
GC% is the percentage of G and C in the primer. It is calculated by dividing the sum of G and C with the total number of bases present in the primer.
Example: Say the primer sequence is ATCGATACGTAG. The GC% of this primer will be (5/12 * 100) = 41.67.
This value is the concentration of primer in nanomoles per unit absorbance (OD) at 260 nm. To calculate extinction coefficient of an oligonucleotide say ATGCA use the following formula -
eATGCA = [2 (eAT + eTG + eGC + eCA) - (eT + eG + eC)]
The extinction coefficient so obtained is in A260 units/µmol. To obtain nmol/A260 multiply the reciprocal of A260 units/µmol by 1000. A260 units/µmol is calculated from table of extinction coefficients given in Appendix A.
Example: Say the primer sequence is ATCGATACGTAG. The extinction coefficient of this primer as calculated from table of extinction coefficients using above formula is 125.9 A260 units/µmol. nmol/A260 for this primer will be 1000/125.9 = 7.94.
This value is the concentration of primer in micrograms per unit absorbance (OD) at 260 nm. To calculate extinction coefficient of an oligonucleotide say ATGCA use the following formula -
eATGCA = [2 (eAT + eTG + eGC + eCA) - (eT + eG + eC)]
This is calculated from table of extinction coefficients given in Appendix A. Then activity in µg/OD is calculated using the following formula:
Activity in µg/OD = Molecular Weight/Extinction Coefficient
Example: Say the primer sequence is ATCGATACGTAG. Its molecular weight will be 3669.47 (as calculated above). Its extinction coefficient will be 125.9. Activity in µg/OD = 3669.47/125.9 = 29.14.
This is the free energy of the primer calculated using the nearest neighbor method of Breslauer, K.J. et. al. DG is calculated by the formula DG = DH - TDS. Here DH is the enthalpy of primer, T is the temperature, DS is the entropy of primer. T is set by DG temp. in the preferences. First the DH and DS are calculated and then the DG is calculated using their values.
Example: Say the primer sequence is ATCGATACGTAG. Its DH and DS will be -85000 cal/mol and -234.7 cal/°K/mol respectively (as calculated below). Its DG will be -85000 - (298.15 * -234.7) = -15024.195 cal/mol = -15.02 kcal/mol.
The stability of the primer determines its false priming efficiency. An ideal primer has a stable 5' end and an unstable 3' end.
If the primer has a stable 3' end, it will bond to a site which is complementary to it other than the target with its 5' end hanging off the edge. It may then lead to secondary bands.
Primers with low stability at the 3' ends function well because the 3' end bonding to false priming sites are too unstable to extend.
The 3' end stability is the DG value of the 5 bases of primer taken from 3' end. The lower this value, numerically, the more liable the primer is to show secondary bands.
Example: Say the primer sequence is ATCGATACGTAG. Its 3' end stability will be DG(CGTAG) = DH(CGTAG) - 298.15 * DS(CGTAG). The DH and DS will be -32200 cal/mol and -82.8 cal/°K/mol resp. Thus its 3' end stability will be -32200 - (298.15 * -82.8) = -7513.18 cal/mol = -7.51 kcal/mol.
This is the enthalpy of the primer as calculated by the nearest neighbor method of Breslauer, K.J. et. al. DH for a pentamer is calculated as follows:
DHATGCA = DHAT + DHTG + DHGC + DHCA
The individual values of DH for nucleotide pairs are taken from the table given in Appendix A.
Example: Say the primer sequence is ATCGATACGTAG. Its DH will be (8600 + 5600 + 11900 + 5600 + 8600 + 6000 + 6500 + 11900 + 6500 + 6000 + 7800) = -85000 cal/mol = -85 kcal/mol.
This is the entropy of the primer as calculated by the nearest neighbor method of Breslauer, K.J. et. al. DS for a pentamer is calculated as follows:
DSATGCA = DSAT + DSTG + DSGC + DSCA
An initiation value of 15.1 is added to the DS calculation. The individual values of DS for nucleotide pairs are taken from the table given in Appendix A.
Example: Say the primer sequence is ATCGATACGTAG. Its DS will be (23.9 + 13.5 + 27.8 + 13.5 + 23.9 + 16.9 + 17.3 + 27.8 + 17.3 + 16.9 + 20.8) + 15.1 = -234.7 cal/°K/mol = -0.23 kcal/°K/mol.
Stability of the 5' termini allows for efficient bonding of the primer to the target site. This stable 5' region is called the GC Clamp. It ensures adequate binding of the primer to the template. Use of primers with optimal stability allows for the use of lower annealing temperatures without the production of secondary bands. Notice that the 3' end should not be very stable and the 5' end should have a strong GC clamp.
The GC Clamp is the DG value of the 5 bases of primer taken from 5' end. The lower this value, numerically, the more efficient is the primer.
Example: Say the primer sequence is ATCGATACGTAG. Its 5' DG will be DG(ATCGA) = DH(ATCGA) - 298.15 * DS(ATCGA). The DH and DS will be -31700 cal/mol and -78.7 cal/°K/mol resp. Thus its 3' end stability will be -31700 - (298.15 * -78.7) = -8235.6 cal/mol = -8.23 kcal/mol.
Repeats and Runs increase the likelihood of false priming. Primers having 3 or more dinucleotide repeats or 3 or more base runs are reported. For example, if the primer contains ATATAT, which is dinucleotide AT repeated three times, the primer is reported.
An important factor to consider in the design of a primer is the presence
A hairpin loop is formed when primer folds back upon itself and is
held in place by intramolecular bonding. Because hairpin loop
formation is an intramolecular reaction, it can occur with as few as
3 consecutive homologous bases. To measure the stability of the
hairpin loop formed, measure the free energy. The free energy of the
loop is based upon the energy of the intramolecular bond and the
energy needed to twist the DNA to form the loop. If this free energy
is greater than 0, the loop is too unstable to interfere with the
reaction. If the free energy is less than 0, the loop could reduce
the efficiency of amplification. The Hairpin Report accessed using
the
![]() button on the window allows you to avoid primers containing secondary
structure.
button on the window allows you to avoid primers containing secondary
structure.
Example: Say the primer sequence is ATCGATATTCGAAGAT. It forms two hairpins. One is 3' end hairpin where the primer folds back upon itself and first and last 3 bases bond together and other is internal hairpin where 2nd to 5th and 9th to 12th bases bond together to form hairpin.
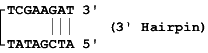
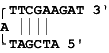
Dimers occur when a region of homology is present within a primer (self -dimer) or between the sense and anti-sense primer (cross dimer). This results in bonding of the two primers, increasing production of the primer dimer artifact and reducing product yields.
Dimers occur within a primer when two copies of the primer bind to each other and cross dimers occur when a primer binds to the other primer in the pair. This is particularly problematic when the homology occurs at the 3' end of either primer. The 3' end will extend readily leading to primer dimer artifact. The dimer and cross dimer reports can be used to test for formation of dimer duplexes.
Example: Say the primer sequence is ATCAGCTGTAGAT. It forms 2 dimers. One is internal dimer where 3rd to 8th bases of primer in 5' to 3' direction (starting from 5' direction) bond with 6th to 11th bases (starting from 3' direction) when primer is placed in reverse direction. The other is 3' end dimer where the last 3 bases (starting from 5' direction) of primer placed in 5' to 3' direction bond with last three base (starting from 3' direction) placed in reverse direction.
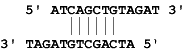
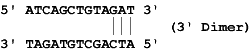
Example: Say the sense primer sequence is ATCAGCTGTAGAT and the anti-sense primer sequence is ATAGTGTAGAT, it forms one cross dimer which is a 3' Cross Dimer.