Home >> Products >> GlycanExplorer >> High-Resolution Accurate-Mass (HRAM) Glycan Spectral Library
High-Resolution Accurate-Mass (HRAM) Glycan Spectral Library
GlycanExplorer accommodates built-in HRAM glycan spectral libraries from various therapeutic glycproteins labeled with fluorophores such as 2-aminobenzamide (2-AB), RapiFluor-MS, Procainamide (ProcA) etc.
The software accommodates spectral libraries of various monoclonal antibody (mAb) variants (Cetuximab, Ipilimumab, Infliximab, Pertuzumab, Golimumab, Nivolumab, and NIST mAb) and other glycoproteins such as Apotransferrin, Orosomucoid, Ovalbumin, RNaseB etc.
| Label | mAb Sample | No. of Glycans |
| Procainamide (ProcA) | Cetuximab | 19 |
| Human IgG | 18 | |
| Ipilimumab | 14 | |
| Nivolumab | 11 | |
| Pertuzumab | 12 | |
| RNase B | 17 | |
| Ovalbumin | 17 | |
| NIST mAb | 17 | |
| Infliximab | 14 | |
| Golimumab | 17 | |
| Fetuin | 17 | |
| Etanercept | 16 | |
| Orosomucoid | 9 | |
| 2-AB | Human IgG | 19 |
| Infliximab | 10 | |
| Etanercept | 13 | |
| RNase B | 15 | |
| Ovalbumin | 23 |
| Label | mAb Sample | No. of Glycans |
| RapiFluor-MS | Human IgG | 30 |
| NIST mAb | 20 | |
| Infliximab | 23 | |
| Golimumab | 26 | |
| Pertuzumab | 15 | |
| RNase B | 15 | |
| Fetuin | 12 | |
| Ovalbumin | 15 | |
| Etanercept | 23 | |
| Cetuximab | 34 | |
| ApoTransferrin | 9 | |
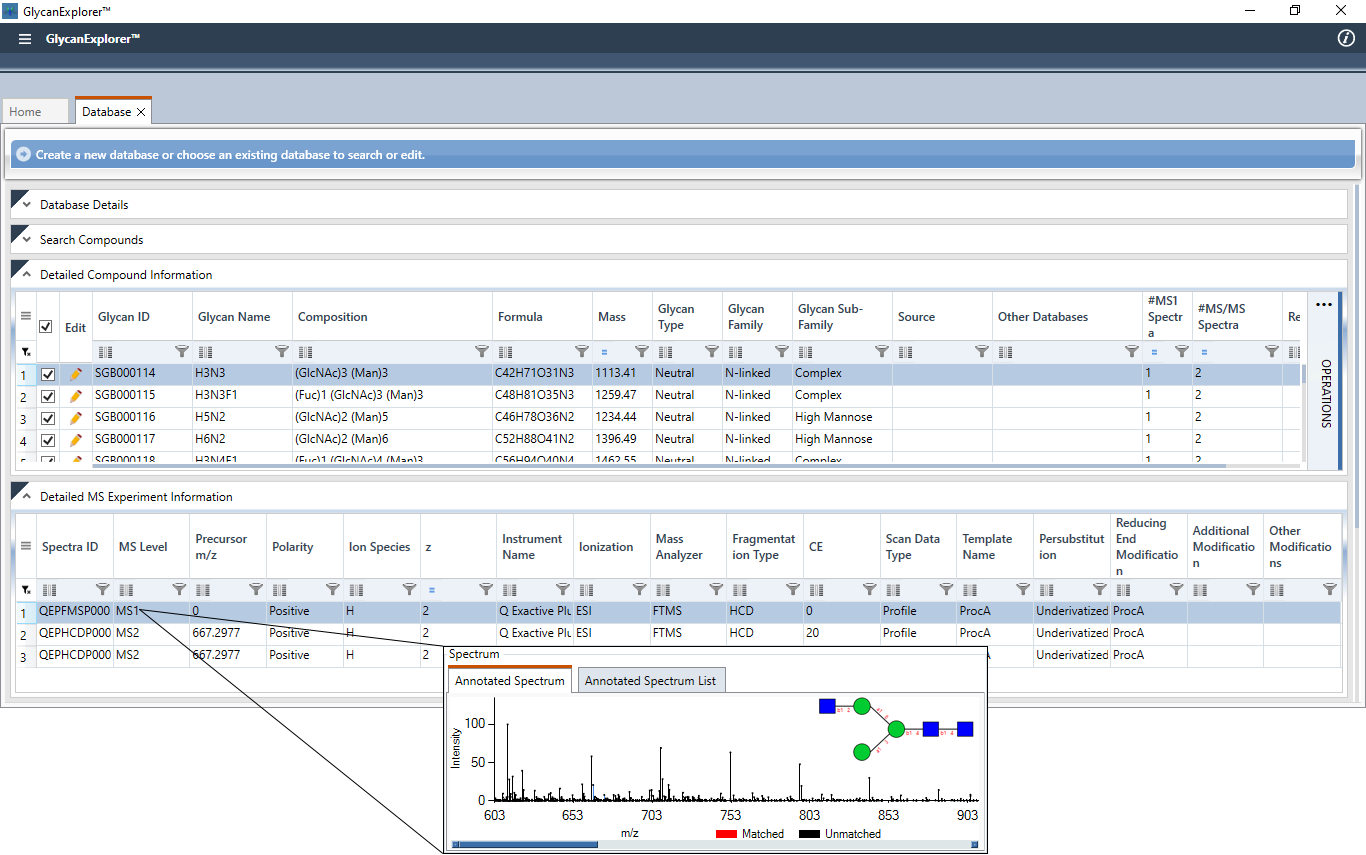
Create a Spectral Library
Apart from the built-in databases, GlycanExplorer allows you to create spectral databases of your glycans of interest. You can import multiple glycan structures, specify their experimental details, store multiple mass spectra (MS1 and MS/MS) and their associated chromatographic information. To help you create a High-Resolution glycan mass spectral library, the software also allows you to calibrate precursor and product ions of an imported mass spectrum based on a single or multiple reference ion(s) of that spectrum. A spectral database can be created following these quick steps:
- Collect KCF (Kegg Chemical Function) contents/ sequences of the constitute glycan structures in a single text file (Sample KCF file, Sample Sequence file).
- Specify the Database name, Description and import the glycan structures using the KCF/Sequence file.
- Specify instrument details, glycan specific experimental details (Ionization technique, Fragmentation technique, Reducing end Fluorophore etc.), and Sample Inlet details (Direct Infusion or Separation).
- Import glycan specific MS1 and MS/MS spectra, specify ion species information, and annotate the fragments of the spectrum by matching with glycan specific in-silico fragments.
- Once successfully annotated, the spectra are saved and further used as reference spectra for reporting candidate glycan structures.
Product Info
Database Statistics
- 14 glycoproteins
- 10000+ MS/MS Spectra
- 200+ MS1 Spectra
Scalable Database
1. Store glycan structures along with retention times, and other experimental and chromatographic details as LC-MS templates.
2. Store multiple spectra of a glycan belonging to different ion species acquired using different instrumental and experimental conditions.
3. Create multiple custom database or LC-MS templates.


