High Throughput Shotgun Lipidomics Analysis Workflow for MALDI-MS Data using SimLipid® Software
August 04, 2020
Lipidomics, an omics discipline, is a study about quantitative characterization of lipids generated by cells, tissues and organisms. Mass spectrometry (MS) plays a vital role in lipidomics enabling accurate identification of lipids of different classes and subclasses with high sensitivity. Shotgun lipidomics, one of the powerful approaches involves a direct high throughput analysis of lipid extracts from various biological sources.
One of the key features of shotgun lipidomics that provides an advantage over the LC based lipidomics is its ability to identify and quantify lipids without any time constraints. To perform shotgun lipidomics, some of the commonly used MS based approaches are direct infusion using electrospray ionization (ESI), matrix-assisted laser desorption ionization (MALDI) based MS, triple quadrupole (QQQ), quadrupole time of flight (QTOF) and TOF/TOF based MS methods.
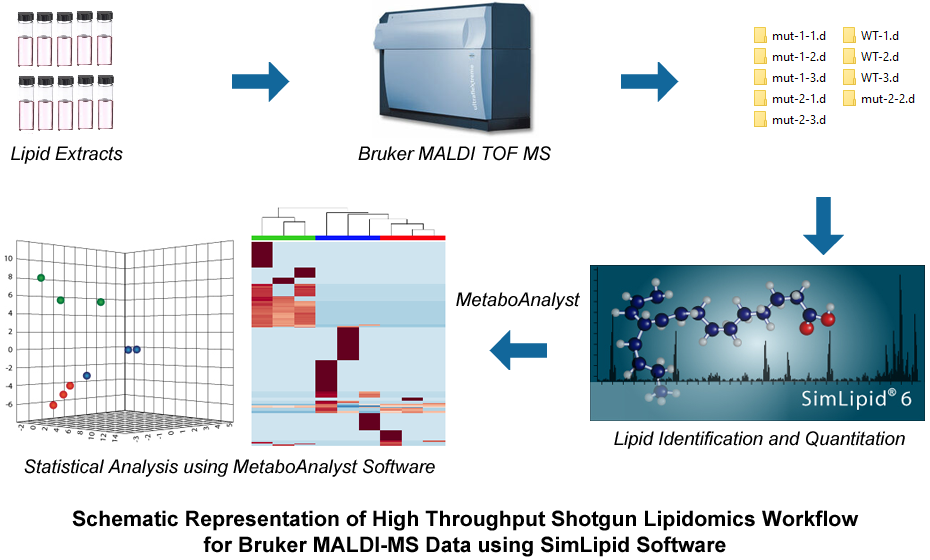
Automated MS data analysis has emerged as the gold standard approach for identification of lipids. SimLipid software offers accurate high-throughput identification capability, and quantitation using robust data processing methods and unique proprietary algorithms. A schematic representation of high throughput shotgun lipidomics workflow using Bruker MALDI-MS data is represented below. Lipid extracts for MS analysis were obtained from a wild type (WT) and two mutant replicate samples (mut-1 and mut-2). Data processing was performed to collate the intensity values of the m/z peaks and later subjected to high throughput MS analysis for lipid identification. During quantitation, the m/z peaks were aligned across spectra of different samples and lipids were annotated. For performing statistical analysis, Metaboanalyst software was used as shown in the workflow.
| Comment | Share |
|


