Home >> Products >> PrimerPlex >> ASPE
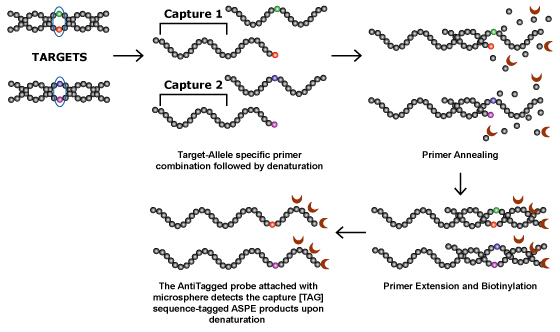
Allele Specific Primer Extension (ASPE) is a solution based, sequence specific enzymatic reaction technology that can be used to assay multiple SNPs in a single tube. The ASPE method involves two phases, an enzymatic reaction that determines the target genotype followed by a capture on solid microsphere surface for detection. Taking advantage of the solution phase kinetics, this technique allows sequence labeled microspheres to be used for detecting new templates. This is done with the help of an appropriate capture sequence attached to the allele specific oligonucleotide.
Design strain differentiation Allele Specific Primer Extension (ASPE) xMAP®assays with AlleleID®.
Allele Specific Primer Design Guidelines
1. ASPE primers should be from the same DNA strand (per target sequence) and be synthesized for all the sequence variants.
2. The Ta opt should be around 51-56 °C.
3. The SNP should be present at the 3' end of the allele specific primer for accurate hybridization.
4. The capture sequence (TAG sequence) should be at the 5' end of the primer.
The template for the ASPE reaction is the amplified PCR fragment. No labeling of primers is done to generate this fragment. In the allele specific primer extension step, polymerases extend the primer by incorporating biotin-labeled dNTPs. Extension only occurs if the 3' end of the allele specific primer is bound to the homologous allelic sequence.
The reaction conditions for this SNP genotyping assay depends upon the anti-TAG sequence that is attached to a fluorescent microsphere. The microspheres are internally labeled with fluorescent dyes and coupled with the anti-TAG sequences.
Allele specific oligonucleotides can be developed by following the assay design guidelines outlined here or by using PrimerPlex, a software tool for designing ASPE assays for multiplex analysis on xMAP based systems.