A High Throughput Qualitative and Quantitative Lipidomics Workflow using FIA-HRMS with Tandem MS (DIA) Based Method with SimLipid Software
September 09, 2019
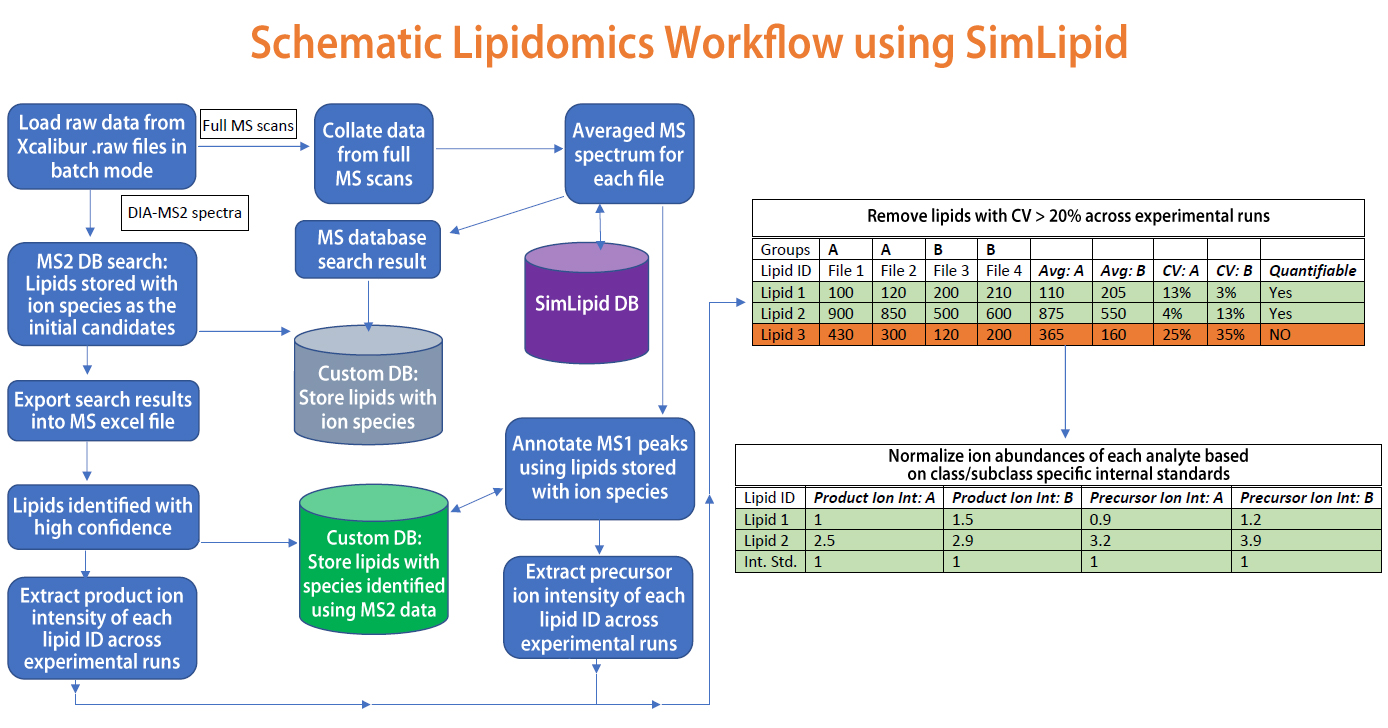
Application of flow injection analysis (FIA) coupled with high-resolution mass spectrometry (HRMS) is currently trending as a suitable method for lipidomics study. In this method, FIA coupled with Thermo Orbitrap MS instrument assists in measuring data-independent acquisition (DIA) based MS/MS data for lipid species. MS and MS/MS data acquired were subjected to data processing, lipid identification, and quantitation using SimLipid software. Averaged MS data were extracted from the raw MS data during loading with a peak m/z tolerance of 5ppm. A selective lipid database search with 5ppm mass tolerance was performed. The results obtained were converted into a custom database (DB MS1) and imported to the SimLipid server database program. The MS/MS data were subjected to a search on the custom database (DB MS1). From these results, the unlikely lipid ion species were removed manually and a total of 592 unique lipids were identified with high confidence. These unique lipids were exported to Microsoft Excel and total ion abundances for the product ions were extracted across the experimental runs. The coefficient of variation (CV) was calculated for each lipid species using in-built Excel formulas and only those lipid ions (330 unique) with CV<20% were selected. The product ion abundances along with their respective parent lipid ion species were stored as another custom database (DB MS2). The averaged MS spectra were re-annotated using the DB MS2 custom database and 206 unique lipids with a CV < 20% of precursor ion abundances across replicates were obtained. The ion abundances of the class-specific internal standard were used for quantitation. Also, a comparative analysis was performed across study groups using normalized precursor versus product ion abundances. Both cases, wherein lipid species that showed similar and different patterns in terms of increase/decrease in normalized precursor ion abundance versus the normalized product ion abundances were compared. For more details, please visit http://premierbiosoft.com/citations/lipidomics-posters-technical-application-notes.html.
Lipidomics Analysis using LC-MS data from Thermo Orbitrap Velos Pro MS instrument with SimLipid Software
August 12, 2019
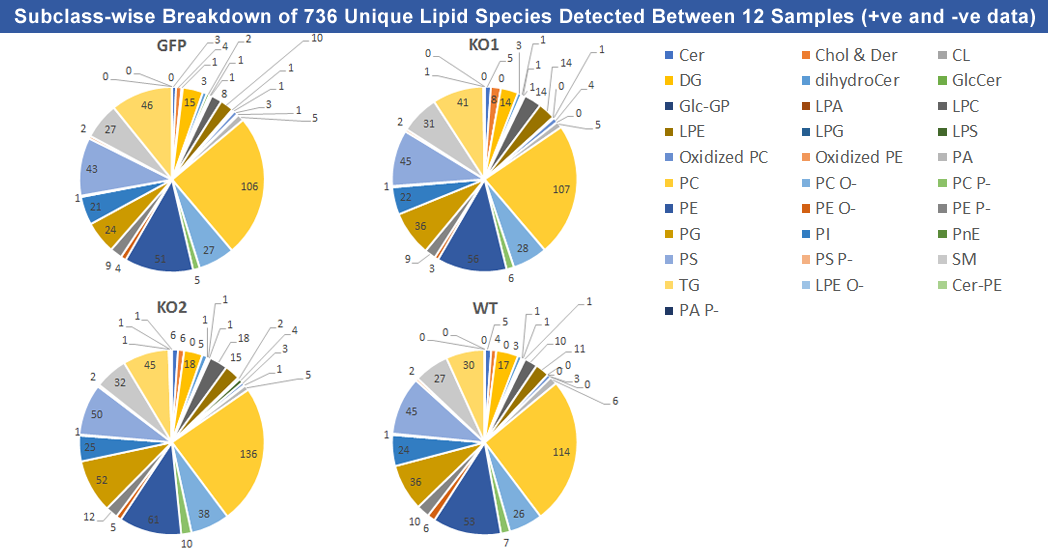
Recent advances in the fields of liquid chromatography (LC) and mass spectrometry (MS) has led to the development of novel lipidomics methods. We have developed a new LC-MS based lipidomics method for lipid profiling of Plasmodium berghei samples using UHPLC coupled with high resolution Orbitrap Velos Pro MS instrument. This lipidomics analysis workflow comprises raw data processing, lipid identification, and statistical analysis. SimLipid v. 6.05 software was used for raw data processing and lipid identification and SIMCA-P (HYPERLINK) software was used for statistical analysis. Lipid identification in SimLipid is based on exact mass database search as well as MS/MS database search of the structure-specific MS/MS characteristic ions using a stringent mass tolerance of 5 ppm for both the precursors and product ions. Comprehensive characterization of lipids – head group, and fatty acyl composition - was achieved using MS/MS data from both positive and negative ion modes. Label free quantitation of the identified lipid species was performed using ion abundance corresponding to its monoisotopic peak of the isotope cluster. During quantitation, one of the challenges observed for detected lipids from LC-MS data is the convoluted LC-MS peaks of isomeric compounds. Hence, we used the total ion abundances of the LC peaks corresponding to isomeric lipid species that have the same number of carbons and double bonds in fatty acid chains. Lipid profiling of P. berghei wild type and hemolysin III knockout strains to identify significant lipid species that discriminate these strains was also performed. A summary of the unique lipid species identified from both positive and negative modes by SimLipid (HYPERLINK) software tool is shown in the figure above. For more details, please visit http://premierbiosoft.com/citations/lipidomics-posters-technical-application-notes.html.
Shotgun Lipidomics of prostate cancer cells using Shimadzu LC/MS 8050 triple quadrupole mass spectrometer and simplified data analysis by SimLipid® software
September 20, 2017
Lipidomics is an emerging field in biomedical research as lipids play an important role in cell, tissue and organ physiology and have potential as biomarkers of disease or treatment success. Lipidomics has also been widely studied in many areas including prostate cancer research, e.g., identification of lipid species in bio-fluids as biomarkers in diagnosis of prostate cancer with high sensitivity.
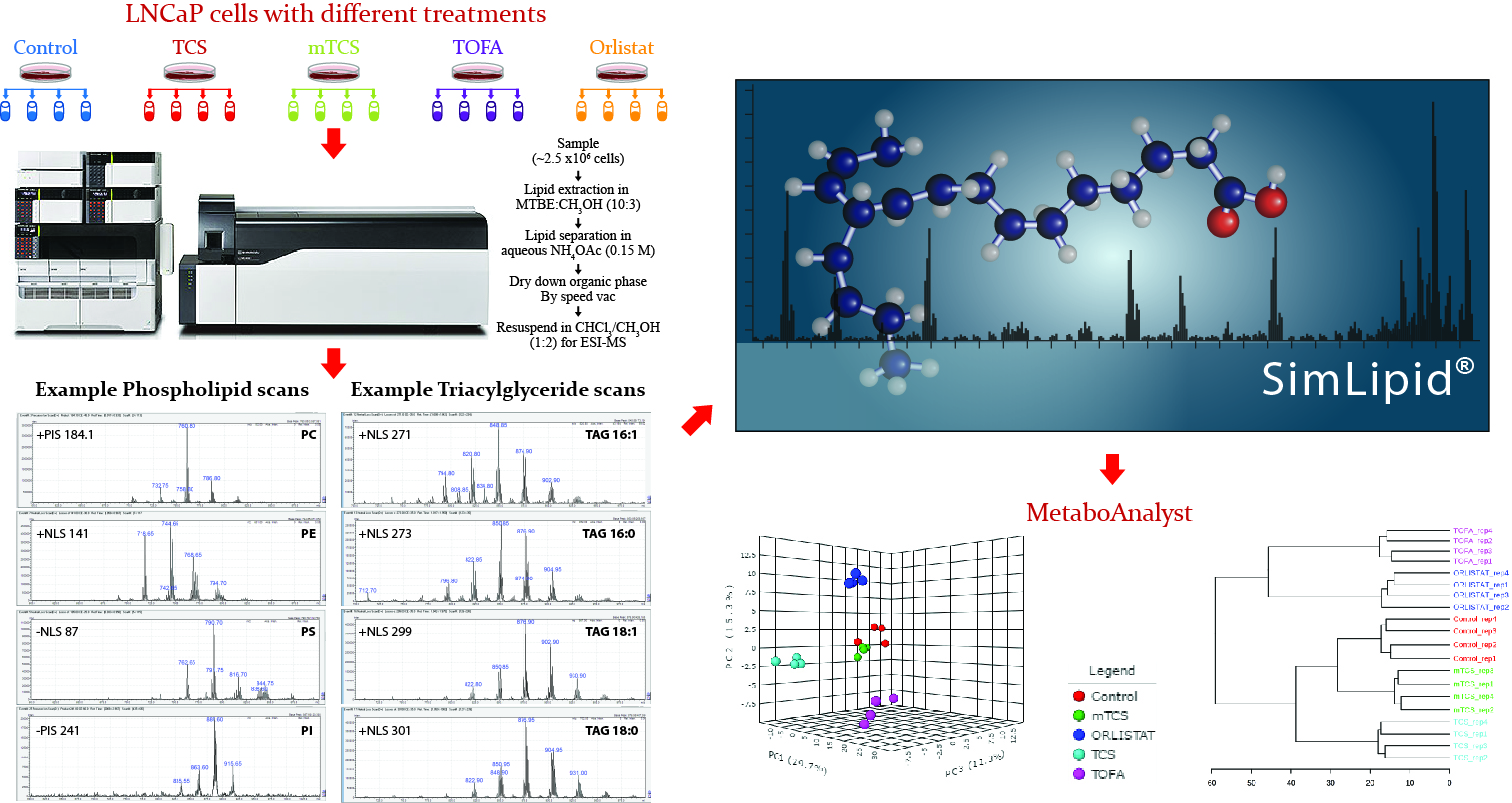
Shotgun lipidomics involves identification and quantification of lipids by direct infusion of complex lipid samples into the mass spectrometer without any chromatographic separation. In this study, we investigate the details of lipid compositions difference between prostate cancer patients treated with different drugs and the control group.
Brief description of the method: ESI based direct infusion method was used for quantitative lipidomics analysis of LNCaP cells with different treatments. Data was acquired following Precursor Ion/Neutral Loss Scanning (PIS/NLS) experiments using Shimadzu LC/MS 8050 triple quadrupole mass spectrometer (http://www.shimadzu.com, Kyoto, Japan). High Throughput data analysis was performed using SimLipid v. 6.0 software (www.premierbiosoft.com, Palo Alto, USA) showing promising functionality for both qualitative and quantitative analysis.
Statistical analysis – hierarchical cluster analysis, and principal component analysis (PCA) - using MetaboAnalyst software was done that clearly separated the study groups and stratified different treatments.


