Application of MALDI MS for Lipidomics
July 13, 2022
Lipids, one of the essential components of cellular membranes, play a critical role in cell-cell interactions like signaling, energy storage, and homeostasis. Unlike proteins, lipid structures are more diverse and complex. Even subtle differences in lipid structures trigger serious biological implications. Thus, there is a need for comprehensive analytical methods to characterize the complex lipid structures. In recent years, mass spectrometry (MS) has emerged as one of the pioneer analytical techniques for structural analysis. Among the different methods of MS, matrix-assisted laser desorption ionization (MALDI) MS has emerged as one of the conventional approaches to achieving fast and accurate characterization of lipids.
Unlike other MS approaches, separation methods are not involved in the MALDI MS approaches. The biological samples are directly loaded onto a matrix and sent into the MALDI instrument for analysis. Earlier, MALDI MS was predominantly used for proteomic studies. However, since the early 2000s, this approach started gaining popularity in lipidomics studies as well. Although conventional, this method suffered two major setbacks - the matrix, and the unavailability of a high throughput data processing solution. One major drawback of selecting a conventional matrix is ion suppression. When irradiated with a laser beam, the matrix itself produces ions, which are likely to mask the original analyte ions. This issue can be resolved by adding various organic bases to the conventional matrices. The use of high-resolution mass spec instruments may nullify the ion interference as well. For efficient data processing and high throughput lipid profiling, we have developed SimLipid® software.
Using SimLipid, you can achieve fast and accurate lipid identification and quantitation by performing an MS search across the vast SimLipid master database. The database is curated with over 40,000 lipid structures corresponding to all nine lipid categories containing over five million structure-specific in-silico MS/MS characteristic ions. SimLipid has a built-in lipid quantitation module that allows comparative analysis of lipids across different samples using one or more internal standards.
SimLipid exports the lipid analysis results in Microsoft Excel/CSV file format and generates interactive charts and tables (see figure: workflow) for quantitative comparison among multiple samples. To help with further downstream analysis, the software also allows you to export the analysis results in a MetaboAnalyst compatible file format.
In a recent MALDI-MS-based experiment, the lipids were extracted from a biological sample that comprised three different types of variants (WT, MT-1, MT-2). A Bruker MALDI-ToF MS instrument was used for the fast and accurate identification of lipids across the three different samples. The analysis was performed in a positive mode with Hydrogen as an adduct and 5 ppm error tolerance for MS search. SimLipid software was able to annotate 230 lipid structures using an MS1-based search. The three variant samples were aligned prior to the database search and later the internal standards were assigned for each class to generate the quantitation report. The bar chart and pie chart were generated for visualization of lipid class percentages. Also, the ion abundance/frequency table was generated. The quantitation report was exported in a .csv format and MetaboAnalyst was used for statistical analysis. For more information, please schedule a meeting with us.
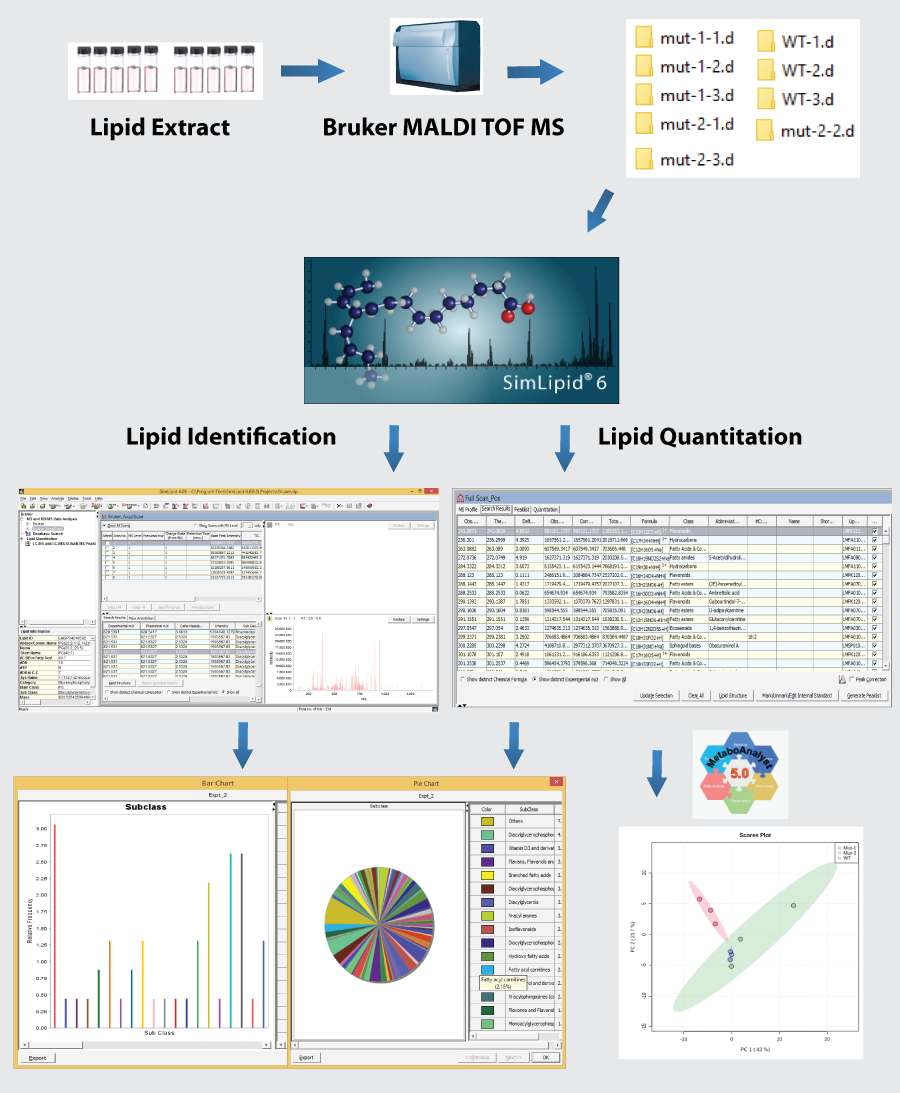
Fig: MALDI MS lipid data analysis workflow in SimLipid.
| Comment | Share |
|


