Robust Lipid Structure Database
SimLipid database is a scalable relational database containing 9 lipid categories namely, Carnitines, Fatty acyls, Glycerolipids, Glycerophospholipids, Polyketides, Prenols, Saccharolipids, Sphingolipids and Sterols. The database stores 40,312 lipids, and 1,508,293 structure-specific in-silico MS/MS characteristic ions. Besides this, the database also stores 5227 transitions of precursor ion scan and neutral loss scan triple quadrupole mass spectrometry methods corresponding to 1,179, 393, and 221 unique fatty acyls from glycerophospholipids, glycerolipids, and sphingolipids respectively.
User can create custom databases* to store curated lipids along with retention times, and drift time. Once a custom database is created, MS, and MS/MS database search for structural identification of lipids can be performed using retention time, and drift time tolerances as initial search predicates. This functionality facilitates users to create LC-MS or LC-Ion Mobility-MS templates; thereby enabling them to exploit the state of the art LC-MS and IMS technologies for their research.
Other database links include Kyoto Encyclopedia of Genes and Genomes (KEGG), Human Metabolome Database (HMDB), Chemical Entities of Biological Interest (CEBI), PubChem Substance database, LIPIDBANK, and LIPIDAT. The database is continuously being updated.
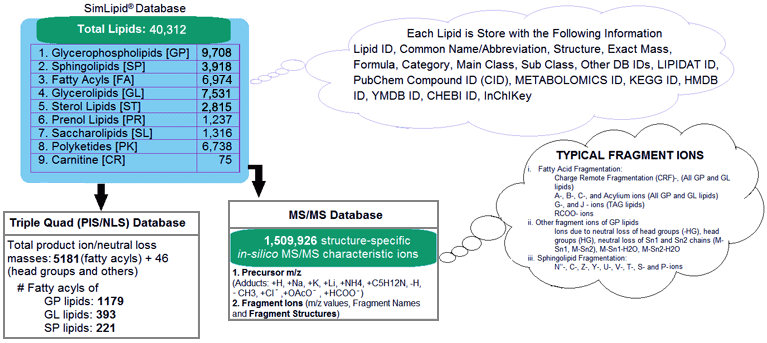
Schematic representation of SimLipid database.
Please Note: * The functionality is available in the Enterprise Edition of the program.


