References >> NASBA Technology
NASBA TechnologyIntroduction to NASBA
RNA detection is commonly done using RT-PCR, a time consuming process often resulting in false positives due to cross contamination. Alternatively, nucleic acid sequence-based amplification (NASBA) is a one step isothermal process for amplifying RNA. NASBA has proven to be successful in detection of both viral and bacterial RNA in clinical samples.
A NASBA reaction consists of avian myeloblastosis virus (AMV), reverse transcriptase (RT), T7 RNA polymerase and RNase H with two oligonucleotide primers. The amplification is more than 1012 fold in 90 to 120 minutes. The amplification of ssRNA is possible only when the denaturation of dsDNA does not occur. As NASBA is an isothermal process, it is thus possible. The NASBA reaction does not get false positives caused by genomic dsDNA, as in the case of RT-PCR.
Mechanism
A schematic representation of the NASBA amplification is shown below:
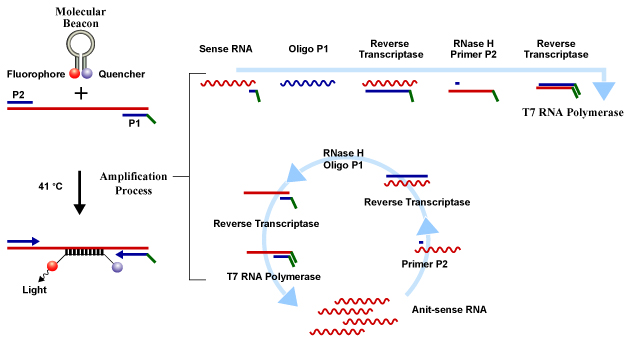
This mechanism requires two short single stranded DNA fragments primers and three enzymes.
-
The viral RNA strands are represented as the sense strand present in the original samples.
-
Primer P1 binds to the RNA and is elongated by reverse transcriptase (AMV-RT). The RNA strand of the yielded DNA : RNA hybrid is hydrolyzed by RNase H.
-
After the binding of P1, primer P2 can also bind. Primer P2 is then elongated by AMV – RT, yielding a double-stranded DNA molecule.
-
Primer P1 is designed in such a manner that when it forms a double- stranded DNA, it codes for a T7 RNA polymerse Promoter site. This helps in generating antisense RNA copies using a DNA template.
-
The new copies of DNA are generated using RNA. The process is same as followed for sense strand. Here in this case, P2 will bind first.
In a NASBA reaction, DNA is the final product formed. It has a promoter region. This allows T7 RNA polymerase to use it as a template. The reaction sequences starting from the sense and antisense RNA strands are different but they are considered to be kinetically identical. They both are referred to as copy DNA (cDNA).
Molecular Beacon in NASBA
A real-time detection system is generated using molecular beacons with the NASBA amplification. Molecular beacons are single stranded hairpin shaped oligonucleotide probes. Their 5' end has a fluorescent label and the quencher is present at 3' end. These molecular beacons are highly specific to their targets. They form a stable hybrid with their amplified target RNA, when present in a NASBA amplification reaction.
Advantages of NASBANASBA is a technique with widespread applications in the area of RNA amplification and detection. The following are the advantages: 1. The amplification of nucleic acid sequence of more than 109 copies can be done in just 90 minutes by the three-enzyme action. |
Software for NASBA
Using Beacon Designer, molecular beacons can be designed for NASBA® assays. Beacon Designer can design molecular beacons for NASBA® assays for studying differential gene expression or for allelic discrimination. You can either design primers for a sequence or also evaluate pre-designed primers and probes for NASBA® assays.


