References >> Gene Splicing
Gene Splicing Overview & TechniquesGene Splicing Introduction
Gene splicing is a post-transcriptional modification in which a single gene can code for multiple proteins. Gene Splicing is done in eukaryotes, prior to mRNA translation, by the differential inclusion or exclusion of regions of pre-mRNA. Gene splicing is an important source of protein diversity. During a typical gene splicing event, the pre-mRNA transcribed from one gene can lead to different mature mRNA molecules that generate multiple functional proteins. Thus, gene splicing enables a single gene to increase its coding capacity, allowing the synthesis of protein isoforms that are structurally and functionally distinct. Gene splicing is observed in high proportion of genes. In human cells, about 40-60% of the genes are known to exhibit alternative splicing.
Gene Splicing |
Gene Splicing Mechanism
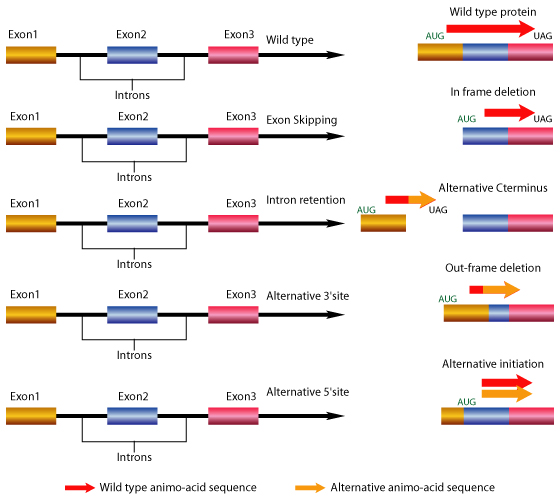
There are several types of common gene splicing events. These are the events that can simultaneously occur in the genes after the mRNA is formed from the transcription step of the central dogma of molecular biology.
Exon Skipping: This is the most common known gene splicing mechanism in which exon(s) are included or excluded from the final gene transcript leading to extended or shortened mRNA variants. The exons are the coding regions of a gene and are responsible for producing proteins that are utilized in various cell types for a number of functions.
Intron Retention: An event in which an intron is retained in the final transcript. In humans 2-5 % of the genes have been reported to retain introns. The gene splicing mechanism retains the non-coding (junk) portions of the gene and leads to a demornity in the protein structure and functionality.
Alternative 3' Splice Site and 5' Splice Site: Alternative gene splicing includes joining of different 5' and 3' splice site. In this kind of gene splicing, two or more alternative 5' splice site compete for joining to two or more alternate 3' splice site.
Splice Variant Detection Methods
Gene splicing leads to the synthesis of alternate proteins that play an important role in the human physiology and disease. Currently, the most efficient methods for large scale detection of splice variants include computational prediction methods and microarray analysis. Microarray based splice variant detection is the most popular method currently in use. The highly parallel and sensitive nature of microarrays make them ideal for monitoring gene expression on a tissue-specific, genome-wide level. Microarray based methods for detecting splice variants provide a robust, scalable platform for high-throughput discovery of alternative gene splicing. A number of novel gene transcripts were detected using microarray based methods that were not detected by ESTs using computational methods. Another commonly used method for discovering of novel gene isoforms is RT-PCR followed by sequencing. This is a powerful approach and can be effectively used for analyzing a small number of genes. However, it only provides only a limited view of the gene structure, is labor-intensive, and does not easily scale to thousands of genes or hundreds of tissues.
Challenges in Microarray Design for Splice Variant Detection
Microarray based gene splicing detection poses some unique challenges in designing probes for isoforms that show a high degree of homology. In order to differentiate between these isoforms, a microarray that uses a combination of probes for exons and exon-exon junctions is used. Exon skipping events or other deletions can be monitored by using junction probes. For example, a probe spanning the exon 1 and exon 3 of the gene will detect the skipping of exon 2 from the gene that is translated into a protein.
Software for Designing Splice Variant Microarrays
AlleleID® automatically designs junction probes as well as intra-exon probes. With AlleleID® you can design any combination of probes to detect any type of alternative splicing event.


