Spectral Matching Vs. in-silico Fragment Matching – Rendering Confidence to Bio-therapeutic Glycan Characterization
January 13, 2021
Recent advances in the field of mass spectrometry has opened new dimensions in biopharmaceutical and biomedical analysis. Monoclonal antibodies (mAbs) are acting as a driving force in the growth of the biopharmaceutical industry. Monosaccharide compositions, glycosidic linkages, and abundances of glycans in the glycoprotein molecules are now important for understanding the efficacy and batch-to-batch consistency of different drugs. This has led to the evolution of tandem MS (MS/MS) technique, which is proven to be more successful compared to the first stage mass spectrometry (Full MS) technique.
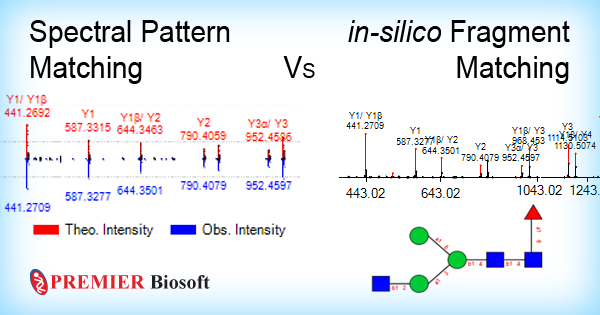
The conventional MS/MS-based identification strategy involves identifying candidate glycan structures based on the number of theoretical fragments (related to the glycan structure) present in the experimental MS/MS spectrum. This technique needs all possible theoretical fragments of a glycan to be available to compare with the experimental fragments. Over time, it was realized that this in-silico fragment matching strategy works well for discovery analysis, however, may not be an ideal approach for the bio-pharmaceutical industry because of the type of the molecules they work with and the workflows they employ. Biopharmas follow a targeted analysis workflow on pre-characterized molecules, and hence adopting a time consuming de novo identification strategy may not be justified.
A new technique, spectral pattern matching addresses these challenges. The process involves creating a spectral library with the help of pre-analyzed glycan standards under a set of instrumental and experimental conditions. The reference spectra contains more precise fragment information compared to in-silico fragments, which makes the identification more confident and time-efficient. Apart from the individual in-silico matching and spectral matching algorithms, certain software products, such as GlycanExplorerTM, are equipped with a unique search mechanism that renders dual confidence to the glycan identification.
| Comment | Share |
|


