References >> Microarray
Microarray Technology How microarray technology works?
|
Bacterial identification using microarrays |
Gene splicing detection using microarrays |
Introduction to Microarray
Molecular Biology research evolves through the development of the technologies used for carrying them out. It is not possible to research on a large number of genes using traditional methods. DNA Microarray is one such technology which enables the researchers to investigate and address issues which were once thought to be non traceable. One can analyze the expression of many genes in a single reaction quickly and in an efficient manner. DNA Microarray technology has empowered the scientific community to understand the fundamental aspects underlining the growth and development of life as well as to explore the genetic causes of anomalies occurring in the functioning of the human body.
A typical microarray experiment involves the hybridization of an mRNA molecule to the DNA template from which it is originated. Many DNA samples are used to construct an array. The amount of mRNA bound to each site on the array indicates the expression level of the various genes. This number may run in thousands. All the data is collected and a profile is generated for gene expression in the cell.
Microarray Technique
An array is an orderly arrangement of samples where matching of known and unknown DNA samples is done based on base pairing rules. An array experiment makes use of common assay systems such as microplates or standard blotting membranes. The sample spot sizes are typically less than 200 microns in diameter usually contain thousands of spots.
Thousands of spotted samples known as probes (with known identity) are immobilized on a solid support (a microscope glass slides or silicon chips or nylon membrane). The spots can be DNA, cDNA, or oligonucleotides. These are used to determine complementary binding of the unknown sequences thus allowing parallel analysis for gene expression and gene discovery. An experiment with a single DNA chip can provide information on thousands of genes simultaneously. An orderly arrangement of the probes on the support is important as the location of each spot on the array is used for the identification of a gene.
Types of Microarrays
Depending upon the kind of immobilized sample used construct arrays and the information fetched, the Microarray experiments can be categorized in three ways:
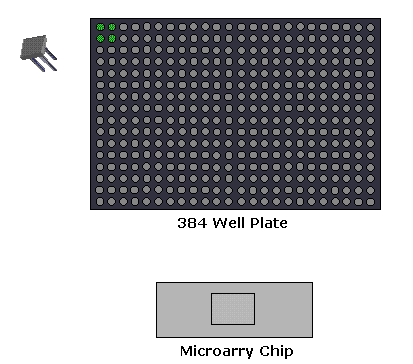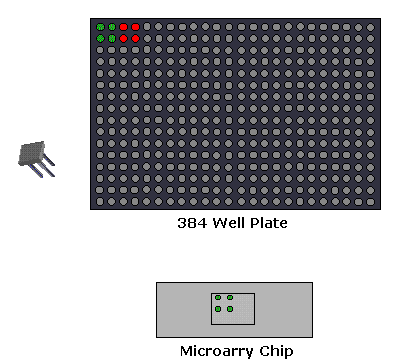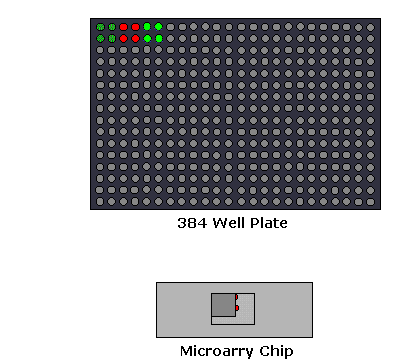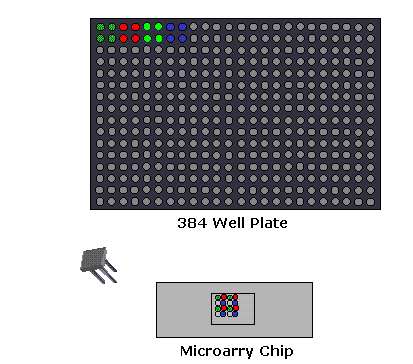 1. Microarray Expression Analysis: In this experimental setup, the
cDNA derived from the mRNA of known
genes is immobilized. The sample has
genes from both the normal as well as
the diseased tissues. Spots with
more intensity are obtained for diseased
tissue gene if the gene is over expressed
in the diseased condition. This expression
pattern is then compared to the expression
pattern of a gene responsible for a
disease.
1. Microarray Expression Analysis: In this experimental setup, the
cDNA derived from the mRNA of known
genes is immobilized. The sample has
genes from both the normal as well as
the diseased tissues. Spots with
more intensity are obtained for diseased
tissue gene if the gene is over expressed
in the diseased condition. This expression
pattern is then compared to the expression
pattern of a gene responsible for a
disease.
2. Microarray for Mutation Analysis: For this analysis, the researchers use gDNA. The genes might differ from each other by as less as a single nucleotide base.
A single base difference between two sequences is known as Single Nucleotide Polymorphism (SNP) and detecting them is known as SNP detection.
3. Comparative Genomic Hybridization: It is used for the identification in the increase or decrease of the important chromosomal fragments harboring genes involved in a disease.
Applications of Microarrays
Gene Discovery: DNA Microarray technology helps in the identification of new genes, know about their functioning and expression levels under different conditions.
Disease Diagnosis: DNA Microarray technology helps researchers learn more about different diseases such as heart diseases, mental illness, infectious disease and especially the study of cancer. Until recently, different types of cancer have been classified on the basis of the organs in which the tumors develop. Now, with the evolution of microarray technology, it will be possible for the researchers to further classify the types of cancer on the basis of the patterns of gene activity in the tumor cells. This will tremendously help the pharmaceutical community to develop more effective drugs as the treatment strategies will be targeted directly to the specific type of cancer.
Drug Discovery: Microarray technology has extensive application in Pharmacogenomics. Pharmacogenomics is the study of correlations between therapeutic responses to drugs and the genetic profiles of the patients. Comparative analysis of the genes from a diseased and a normal cell will help the identification of the biochemical constitution of the proteins synthesized by the diseased genes. The researchers can use this information to synthesize drugs which combat with these proteins and reduce their effect.
Toxicological Research: Microarray technology provides a robust platform for the research of the impact of toxins on the cells and their passing on to the progeny. Toxicogenomics establishes correlation between responses to toxicants and the changes in the genetic profiles of the cells exposed to such toxicants.
GEO
In the recent past, microarray technology has been extensively used by the scientific community. Consequently, over the years, there has been a lot of generation of data related to gene expression. This data is scattered and is not easily available for public use. For easing the accessibility to this data, the National Center for Biotechnology Information (NCBI) has formulated the Gene Expression Omnibus or GEO. It is a data repository facility which includes data on gene expression from varied sources.
Microarray probe design parameters
For 25-35 mers
| Parameter | Minimum Value | Maximum Value | Default Value | Unit |
| Probe Length | 10 |
99 |
30 |
bases |
| Probe Length tolerance | 0 |
15 |
3 |
|
| Probe Target Tm | 40 |
99 |
63 |
°C |
| Probe Tm Tolerance (+) | 0.1 |
99 |
5 |
|
| Hairpin Max ÄG | 0.1 |
99.9 |
4 |
Kcal/mol |
| Self Dimer ÄG | 0.1 |
99.9 |
7 |
Kcal/mol |
| Run/Repeat | 2 |
99 |
4 |
bases |
For 35-45 mers
| Parameter | Minimum Value | Maximum Value | Default Value | Unit |
| Probe Length | 10 |
99 |
40 |
bases |
| Probe Length tolerance | 0 |
15 |
3 |
|
| Probe Target Tm | 40 |
99 |
70 |
°C |
| Probe Tm Tolerance (+) | 0.1 |
99 |
5 |
|
| Hairpin Max ÄG | 0.1 |
99.9 |
6 |
Kcal/mol |
| Self Dimer ÄG | 0.1 |
99.9 |
8 |
Kcal/mol |
| Run/Repeat | 2 |
99 |
5 |
bases |
For 65-75 mers
| Parameter | Minimum Value | Maximum Value | Default Value | Unit |
| Probe Length | 10 |
99 |
70 |
bases |
| Probe Length tolerance | 0 |
15 |
3 |
|
| Probe Target Tm | 40 |
99 |
75 |
°C |
Probe Tm Tolerance
(+/- above) |
0.1 |
99 |
5 |
|
| Hairpin Max ÄG | 0.1 |
99.9 |
6 |
Kcal/mol |
| Self Dimer ÄG | 0.1 |
99.9 |
8 |
Kcal/mol |
| Run/Repeat | 2 |
99 |
6 |
bases |
Other Parameters
-
Probe Location
1. 3' end bias: The oligos chosen should be towards the 3' end of the gene i.e. Default : 3' end.
2. The oligos should be designed by default within 999 bases of 3' end. The range can be from 0 to 1500 bases.
-
The oligos should be free of cross homology (i.e They should be BLAST searched against the appropriate genome category).



