Home >> Products >> AlleleID® >> SYBR® Green Primer Design
Primer Design for SYBR® Green Assays
-
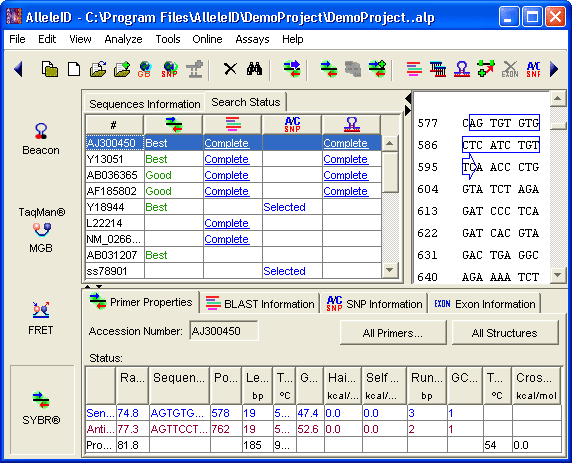
Design Primers: Designs primers optimized for SYBR® Green assays.
-
Avoid Homology: AlleleID® carries out highly specific SYBR® Green primer design by BLAST searching your sequences, automatically interpreting the results and then designing highly specific SYBR® Green primer pairs.
-
BLAST: SYBR® Green primers can be BLASTed against genomic databases available at NCBI. This helps verify the SYBR® Green primer design and visualize primer and amplicon specificity.
-
Template Secondary Structures: Template structures identified are avoided while designing SYBR® Green primers.
-
Algorithm: Calculates SYBR® Green primer Tm using nearest neighbor thermodynamic algorithm.
-
SYBR® Green Primer Rating: Ranks the designed SYBR® Green primers according to expected priming efficiency.
-
Comprehensive Criteria: Screens SYBR® Green primers for their thermodynamic properties, location and secondary structures.
-
Multiplex Primers: Avoids cross homologies with all other dimers to reduce primer dimer.
-
Pre-designed Primers: Designs optimal TaqMan® probes and molecular beacons and for use with well proven SYBR® Green primers. Additionally, you can use a pre-designed or published or well proven forward or reverse primer and have AlleleID® design the rest of the assay. For example, for a given forward primer, AlleleID® would design the reverse primer for SYB®R Green assays and would design both the reverse primer and the TaqMan® probe for TaqMan® assays.
- SNP Amplification: Designs SYBR® Green primers to amplify SNP sites. AlleleID® can even analyze SNP specific primers to help you decide their suitability for the reaction.