Home >> Products >> Primer Premier >> Features
Features
New Improved Search Algorithms
- New improved search algorithms.
- Automatic homology and template structure avoidance.
- Sequence view to visualize primers on the sequence.
- New improved graphical user interface.
Automatic Sequence Retrieval
Quickly retrieves batches of sequences directly into the program from GenBank and dbSNP using accession/GI numbers and assay Ids. Using the multiple retrieval facility, sequences can be loaded from local drives.
Automatic Homology & Template Structure Avoidance
Primer specificity can be achieved by performing BLAST search and search for template structures from within the program. The results of both these searches are used while designing primers. The regions that exhibit significant cross homologies and template structures are avoided automatically during primer design.
Evaluate Pre-designed Primers
Primer Premier analyzes and evaluates the properties of pre-designed primers and ranks them for you to adjudge how far the primers are from their desired target values. This feature is especially useful if you are working with published or pre-designed well proven primers. Primers with up to 5 mismatches can also be evaluated.
For a particular primer, Primer Premier can even design a compatible set. For example, for a pre-designed or published forward primer, Primer Premier can find a compatible reverse primer that meets all the design criteria.
You can evaluate a forward/reverse primer or a primer pair without even having to enter the template sequence. Primer Premier displays all the properties of the oligos being analyzed and exports them in an HTML or csv file format. The primers are ranked enabling you to judge how closely they meet the target values.
Manual Primer Search
Primer Premier offers complete control over primer design in the hands of the user. Using the manual primer search option, a user can click any nucleotide on the template sequence to get primers at the location of their choice. Primer Premier designs a primer with its 3' end located at that nucleotide and analyzes its properties instantly. The length of the primer can also be increased/decreased, per user convenience. For mutagenesis and cloning, this functionality helps in getting an optimal design in an otherwise constrained region. Primer Premier enables "sliding" the primer sequence along the DNA sequence, displaying the properties in real time, enabling users to visualize the best position to locate oligos.
Edit Template Sequence
The template sequence can be edited. Bases can be modified using the standard Cut/Copy/Paste functions, to zero-in on the location of interest. Unwanted regions of sequences can be removed by selecting them and clicking Delete. The program also allows a user to add comments or annotations for the sequence for future reference.
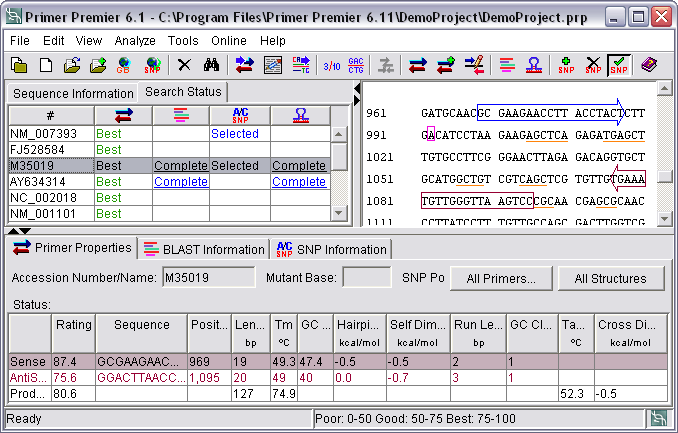
Sequence View
The selected sequence can be viewed in 3 or 10 bases per block, single stranded or double stranded. The designed primers are marked, the sense primer in blue and the antisense primer in red. The selected SNPs are marked in pink. The homologies regions are highlighted in yellow and the bases participating in template structures are underlined. You can export the "sequence view" along with the primer design results in csv or HTML format.
Generate Report
You can create a presentation quality report for the assays you designed for lab notebooks and for sharing information with colleagues. The report visualizes the positions of the primers on the sequence, includes a list of the alternate primers, displays primers, amplicon and sequence properties and the design parameters used by the primer design program.
Local & Desktop BLAST Options
You can BLAST your sequences against local custom databases using the Local and Desktop BLAST options. Create a database of sequences in text or FASTA format on your computer and BLAST your query sequence from Primer Premier. The results are then used for primer design. This support is especially useful if you do not want to submit your data on the Internet or if you are working with unpublished information.
Calculator
A built in calculator calculates the GC%, displays the base count and enables you to obtain the complementary and the reverse complementary strands.
Data & Database Management
Multiple projects can be created. Data of multiple experiments can be easily managed by creating separate project for each experiment. Maintains a local database for sequence information and search results.


