Mass Spectra Interpretation
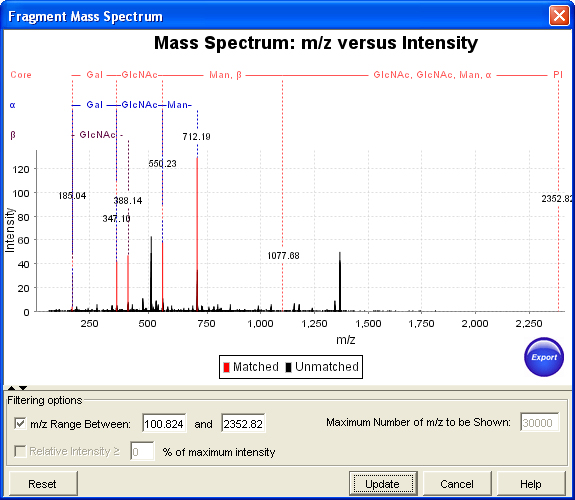
SimGlycan helps in interpreting a mass spectra by highlighting the experimental m/z values that match those of theoretical fragments and generates an annotated spectrum depicting the loss of consecutive monosaccharide units. This helps researcher to establish the branching pattern and verifying the basic unit of glycan. Each peak can be represented using cartoons or Domon Costello nomenclature. Charge states of fragments are also displayed if the fragments are multiply charged.
SimGlycan enables generating an annotated mass spectra either to fit on a page or based on the area of interest. It also exports it for sharing the findings with your colleagues. The simple interface enables you to zoom into specific plot locations, specify m/z range to display specific plot sections and also to export plots as image files to MS PowerPoint, for publishing results and sharing information with colleagues.
Cartoons are constructed using standard Consortium for Functional Glycomics (CFG) nomenclature for monosaccharides (custom symbols are used where CFG annotations are not available).
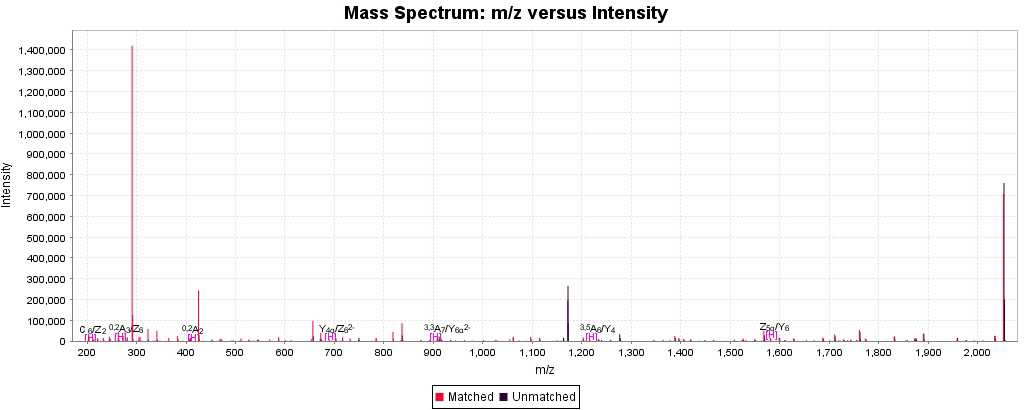
Annotation of the spectra using Domon-Costello fragment names allows the user to see the matched peaks over the spectrum, where glycan fragments are annotated as per standard Domon and Costello nomenclature and peptide fragments are annotated as per its standard names.
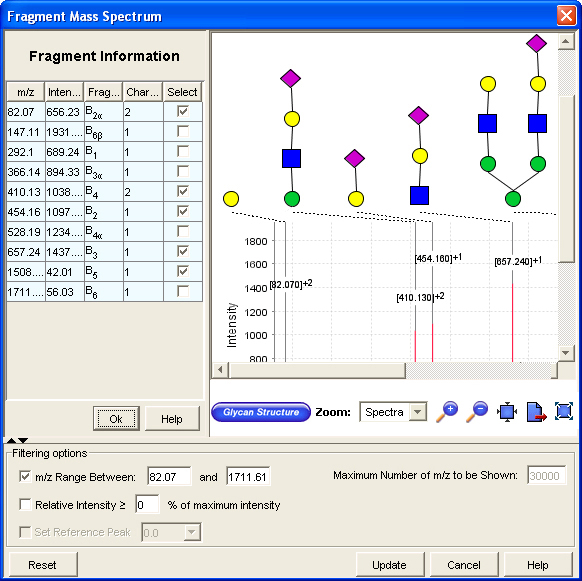
When you launch the Fragment window, the name of the fragment along with its corresponding glycan fragment structure is displayed. The portion in green displays the accepted or active fragment for which the m/z value and mass are calculated and the portion in gray shows inactive or rejected part of the glycan.
Name: Displays the recommended and alternative names of the glycan.
Sequence: Displays the sequence of monosaccharides.
Composition: Displays the glycan composition.
Glycan Mass: Displays the mass computed from the glycan structure.
Precursor Ion m/z: Displays the m/z ratio computed from the composition.
Class: Displays an incomplete but ongoing classification of glycans.
Reaction: Displays biochemical reactions where the glycan appears as a substrate or a product.
Pathway: Displays biochemical pathway maps containing the participating glycans.
Enzyme: Provides information and EC number of enzymes for which the glycan is a substrate or a product.
Other Database Links: Displays all CarbBank entries with the same glycan structure.
Generate Report
You can create a comprehensive glycan analysis report for your lab notebook, publication or sharing information with your colleagues. The report includes the glycan characteristics such as the ID, sequence, name, composition, mass, precursor ion m/z, class, reaction, pathway along with a graphical view of the glycan and its possible theoretical fragments.
SimGlycan highlights the m/z values which match those of theoretical fragments. Upon their selection, it plots, annotates, and exports an MS profile in .png format for your lab notebook. Using a simple interface, you can switch between Column and Scatter plots, zoom into specific plot regions, specify m/z range to display specific plot sections.
SimGlycan enables you to assign your own rank to predicted glycan structures, add comments and search fragments by their mass, name and m/z values.
*Note: Any anion or cation that combines with glycan/glycan fragments to form precursor/product ions. The current version of the program includes comprehensive support for Li and K adducts and adduct combinations such as Na+H, Li+H etc., in addition to the previously supported H and Na adducts.
*Note: The mass spectrometry terms used by us on the web pages and in the software follow the terms and definitions specified in the Standard IUPAC guidelines as described in Murray et al (2006).


