Home >> Products >> AlleleID® >> Allele Specific Oligonucleotide Design
Allele Specific Oligonucleotide Design
Rapid advances in technologies that detect genetic polymorphisms have sped up the discovery of disease related genes and novel tumor markers. The use of DNA microarrays and real time PCR is increasingly becoming widespread. For example, discriminating alleles using a microarray allows simultaneous analysis of several SNPs from a target population in a single experiment.
An allele identification real time PCR assay or microarray detects the alternative forms of a gene. Alternative forms exists due to substitutions, insertions or deletions. Both these techniques involve the use of primers and probes specific to an allele. A wild and mutant type probe are designed for each target SNP such that the SNP is near the center of the probe. Primers are then designed to amplify the region containing the SNP.
Performing Allele Specific Detection with AlleleID®
AlleleID® can design a microarray and/or a real time PCR assay for allele discrimination. To start with, AlleleID® can download sequences directly from NCBI's dbSNP. The SNP information for the sequences is displayed in the SNP information tab. You can also add SNP information to the sequences already downloaded using the Add SNP option.
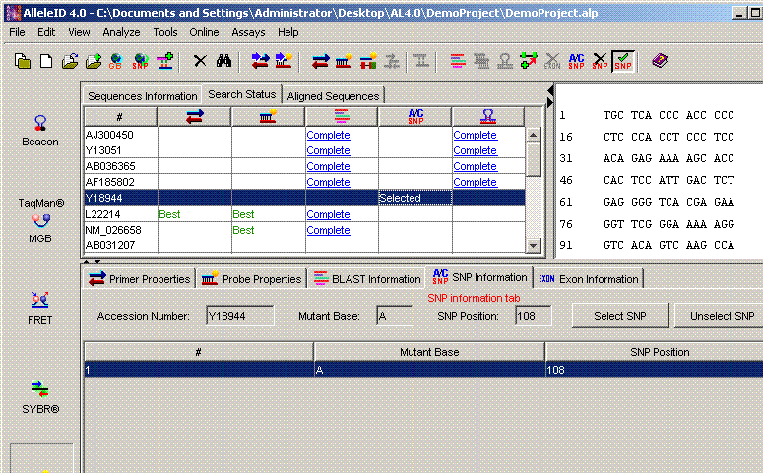
Allele Specific Discrimination Using AlleleID®
AlleleID® designs both the wild and mutant probes for allele discrimination assays for microarrays. It provides provides the following two options:
SNP at the Center of Probe: The probes are designed with the SNP near the center of the probe. This option is used to design probes for allele-specific oligonucleotide hybridization assays or tiling arrays.
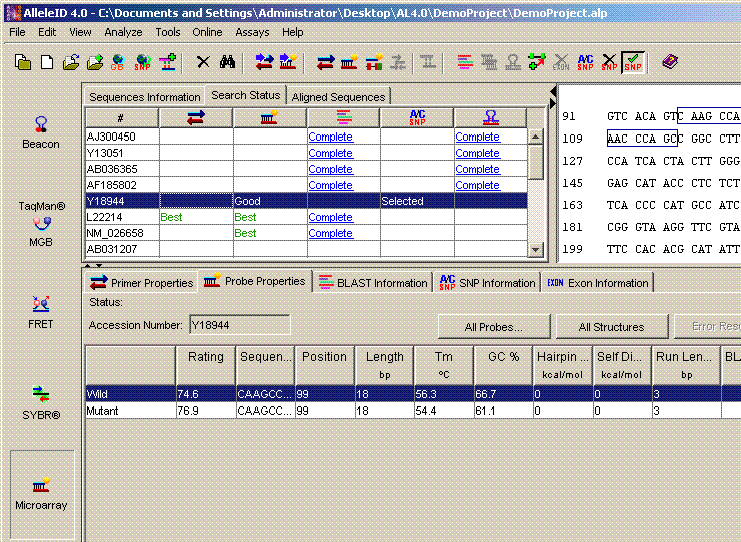
3' End One Base Upstream of SNP Location: Sense and anti-sense probes with their 3' ends one base upstream of the SNP are designed. This option can be used to design probes for allele discrimination using primer extension assays.
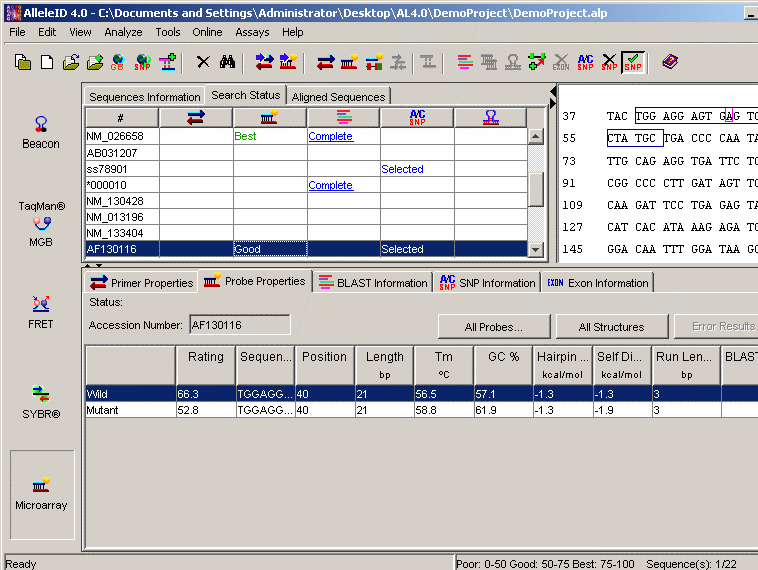
A Real Time PCR Based Allele Discrimination Assay
AlleleID® designs optimal SYBR® Green primers, TaqMan®/TaqMan® MGB probes, FRET probes or molecular beacons for SNP genotyping assays. When a sequence has SNP selected, AlleleID® will design both wild and mutant probes for the sequence. In the SYBR® Green mode, primers are designed so that the nucleotide that distinguishes the alleles is at the 3' terminal base of one of the primers.
For TaqMan®/MGB probes, FRET or molecular beacons assay, probes are designed so that the nucleotide that distinguishes the alleles is located at or near to center of the probe.


