Home >> Products >> AlleleID® >> Species Identification Assays
Species Identification
Using AlleleID®, you can design assays for identifying species and strains from a mix of sequences
- Specifying Sequences: Specify the sequences you would like to detect in the mix by moving them to the "Bind To" box.
- Choices for Primer Design: There are two choices available for primer design, Unique and Optimal. When you choose Unique, the program designs a primer pair to amplify each sequence. For Optimal, the program minimizes the number of primers required to amplify all sequences. The goal is to minimize the synthesis and overall assay costs.
- Choices for Probe Design: Similarly for probes, when you choose Unique, AlleleID® designs one probe per sequence. When you choose the Minimal set option, AlleleID® designs the smallest number of probes required to identify the sequences.
How Species Were Identified Historically
The field of disease diagnosis and drug development has seen a rapid replacement of culture based methods for bacterial and species identification by faster and more reliable molecular based techniques such as real time PCR and microarrays. The reason for the success for these techniques is the ease with which one can identify a species from a sample collected from complex sources such as air, soil, or water.
Molecular based techniques use species-specific probes, designed for specific regions of the target gene, for the detection and identification of bacteria. Microarrays can rapidly detect and identify multiple pathogens simultaneously in complex backgrounds.
Modern Techniques for Species Identification
AlleleID® designs both real time PCR assays, including molecular beacons, TaqMan® probes, TaqMan® MGB, and SYBR® Green primers), and microarrays for bacterial identification, pathogen detection and taxa/species identification. AlleleID® uses the fast and accurate ClustalW algorithm to align sequences to identify the species specific regions.
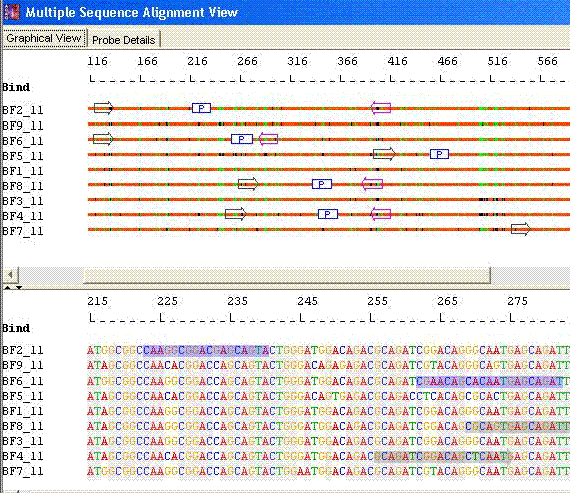
Species Identification: Align sequences and identify unique regions
AlleleID® Provides a Number of Options for Species Identification
- Unique
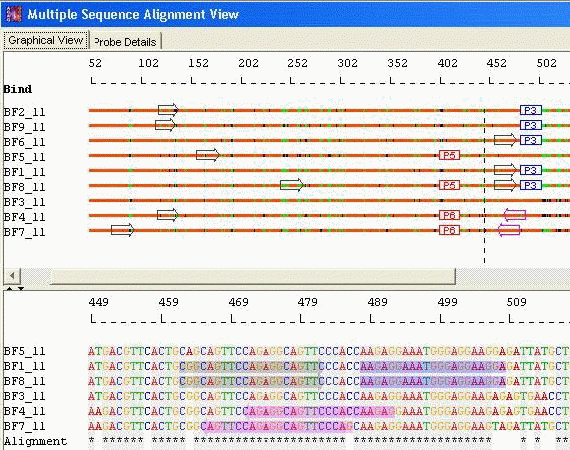
Probes with Unique Primers: When
the probe type chosen is Unique, AlleleID® designs probes to
identify each sequence in the Bind To box, one probe per
sequence. The program
searches for base positions that are unique to each individual
sequence in the alignment. The probes are centered as close as
possible over these unique bases, so that even when the mismatch is
only one base, the assay succeeds.
Species Identification: Unique Probes with Unique Primers
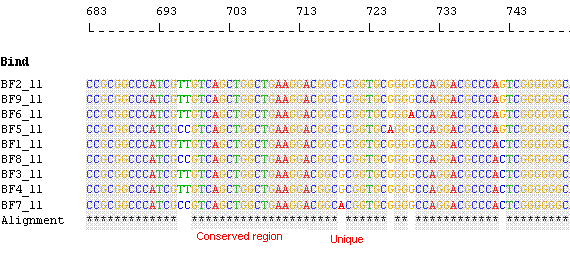
- Minimal
Set with Unique Primers: Choosing the Minimal set option, AlleleID® designs the smallest number of
probes required to identify the sequences. The Minimal set will
generally contain fewer probes than the number of sequences to be
identified, and a unique combination of probes identifies each
sequence. The aim of such an assay is to minimize the
synthesis and overall assay costs.
Species Identification: Minimal Set with Unique Primers
A "Minimal Probe" Matrix
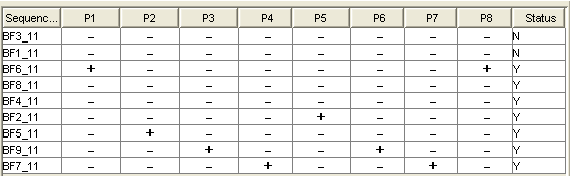
Y stands for the sequences that can be identified uniquely.
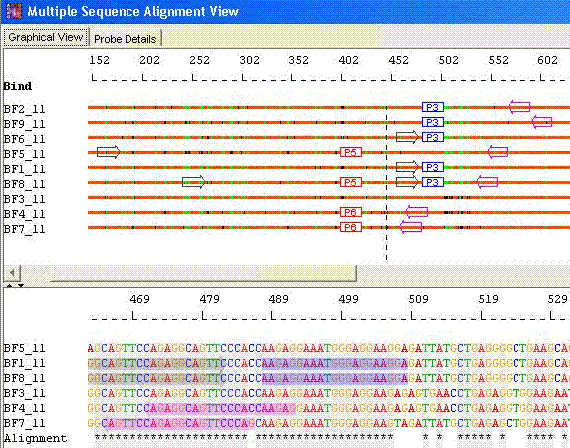
- Minimal Probes with Optimal Primers
For Optimal, the program minimizes the number of primers required to amplify all sequences. The goal is to minimize the synthesis and overall assay costs.
Species Identification: Minimal probes with Optimal primers