Home >> Products >> AlleleID® >> Cross Species Assays
Real Time PCR & Microarray Primer/Probe Design for Cross Species and Taxa Specific Applications
-
For cross species assays, AlleleID® identifies the conserved regions to design a universal probe. This functionality is especially useful to detect a species or a taxa when designing a diagnostic assay. This functionality is also useful to help study gene expression when genome draft of the organism under study is not available.
-
When it is not possible to identify a significant conserved region for a set of sequences, AlleleID® attempts to design a "Mismatch Tolerant" probe. The probe may include up to the specified number of mismatched bases which are located as close as possible to the 5' end.
- Also included is the "Minimal Set" option that helps you design minimum number of probe sets that uniquely identify a sequence reducing the overall assay cost.
Cross Species Detection
Changes in the DNA makeup of the organisms such as single nucleotide polymorphisms, during the course of evolution, have led to the diversity in phenotypic traits. Generations after generations, inherit these changes and contribute towards the biodiversity of species. Despite all these changes, there are still stretches of DNA which remain conserved during evolution. Such conserved regions or sequences are useful in suggesting phylogenetically similar groups, identification of an organism within a group or understanding the extent of its biodiversity amongst the flora of a particular area.
Modern Techniques for Cross Species Detection
Real time PCR and microarrays offer a rapid, sensitive, specific and quantitative tool for the diagnosis of pathogens. Diseases are not readily distinguished based on symptoms when there is more than one pathogen in the sample. Microscopic identification requires culturing on specialized media for days to weeks. Recently, real time PCR assays were successfully used in the detection of fungal pathogens such as Rhizoctonia, Pythium, and Fusarium.
Designing a cross species assay involves the use of a multiple sequence alignment tool to identify conserved stretches of DNA and the design of a hybridization probe for detecting it. Recently there have been advances in micro-organism identification at the taxonomical level using molecular level identification techniques such as real time PCR and microarrays.
How AlleleID® Designs a Cross Species/Taxa Specific Assay
AlleleID® designs both real time PCR assays, including molecular beacons, TaqMan® probes, TaqMan® MGB, and SYBR® Green primers, and microarrays for designing a universal probe for taxa discrimination.
When designing a taxa specific assay, AlleleID® designs a single common probe to identify a taxa or a group of organisms from the mix. For challenging problems when such designs are not feasible, AlleleID® attempts the design by minimizing mismatches, avoiding them especially towards the more problematic 3' end.

AlleleID® Provides Several Design Options for Designing Taxa Specific/Cross Species Assay
Conserved Probes with Conserved Primers: AlleleID® identifies conserved regions by aligning sequences and then designs a probe within them. This enables identification of any sequence in the set with a single probe. It also designs primer pairs in the conserved regions flanking the probes. As a result, a single primer pair can amplify all the sequences of a group or a taxa.
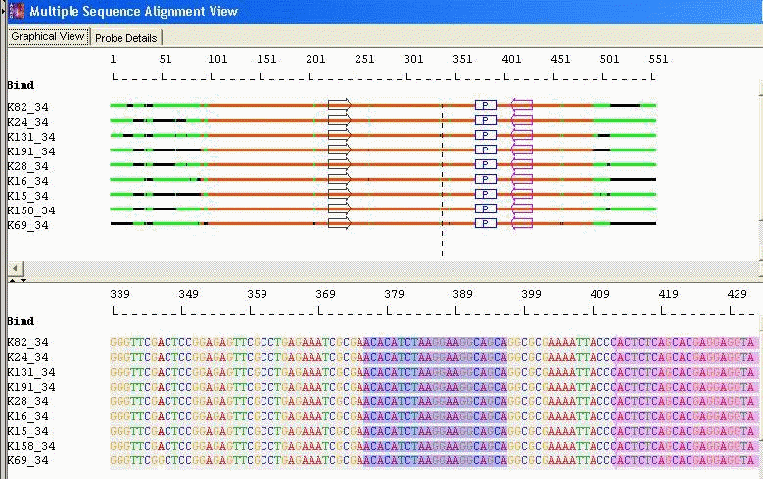
Taxa Specific/Cross Species Identification: Conserved Probe with Optimal Primers
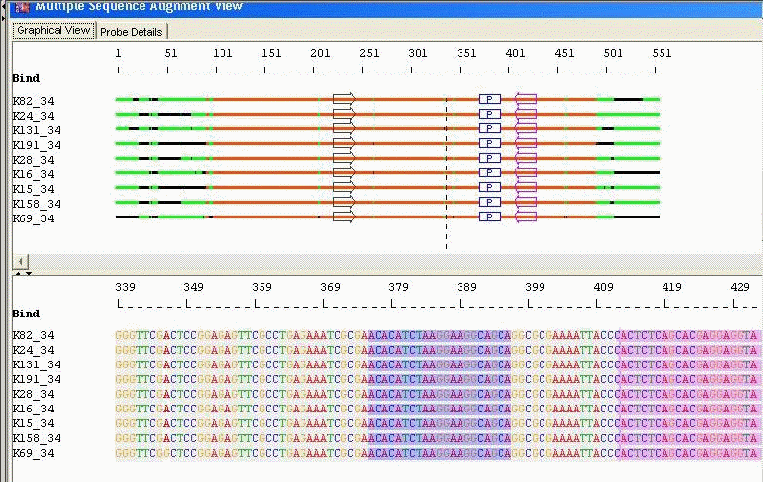
Conserved Probe with Optimal Primers: AlleleID® designs a primer pair that would amplify the maximum number of sequences possible in an alignment. There are two choices available for designing primers, Unique and Optimal. For "Unique", the program designs a primer pair to amplify each sequence. Each primer is completely homologous to its template. For "Optimal', the program minimizes the number of primers required to amplify all sequences; taking advantage of the fact that primers can bind to a sequence and amplify it even if they contain a limited number of mismatched bases especially towards the 5' end of the primer.
Taxa Specific/Cross Species Identification: Probes with Mismatched Bases and Conserved Primer
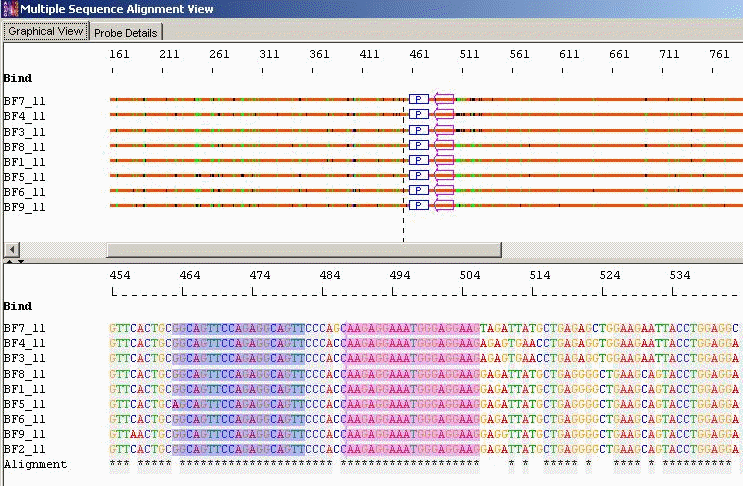
Probes with Mismatched Bases and Conserved/Optimal Primers: AlleleID® can design probes with specified number of mismatch bases. The mismatched bases will follow the majority consensus, meaning that the probe will have the most common base within the alignment. This design minimizes cost, an important consideration, especially in diagnostic assay design. The primers are designed in the conserved regions. To reduce assay costs, it designs the minimal number of primer pairs that are required to amplify the conserved regions across the alignment.
Taxa Specific/Cross Species identification: Probes with Mismatched Bases and Optimal Primers
Minimal Probes with Optimal Primers: When the homology among the aligned sequences is very low and both the designs mentioned above (a single taxa specific probe set or probe set with mismatch tolerance) are not feasible, AlleleID® designs the minimum number of probes possible to detect all the sequences in an alignment. The program designs probes for the majority consensus first and then for the minority consensus and remaining individual sequences. This design is very useful, especially for studying phylogenetic relationships among sequences obtained from different geographical locations.
Taxa Specific/Cross Species Identification: Probes with Mismatched Bases and Conserved Primer
Taxa Specific/Cross Species identification: Probes with Mismatched Bases and Optimal Primers
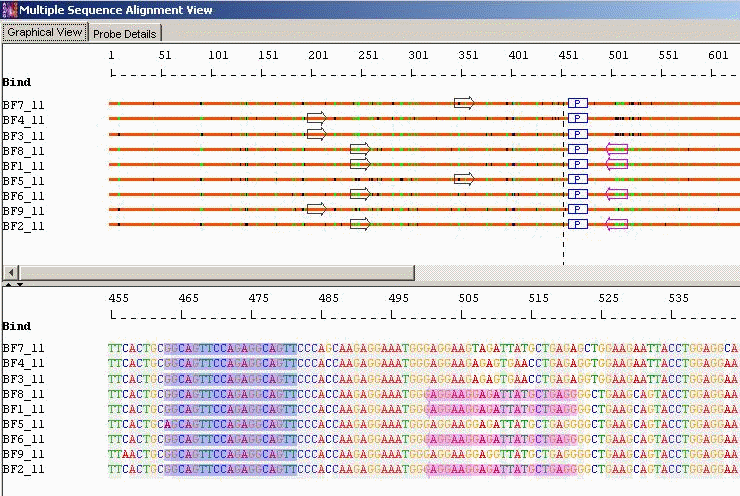
Minimal Probes with Optimal Primers: When the homology among the aligned sequences is very low and both the designs mentioned above (a single taxa specific probe set or probe set with mismatch tolerance) are not feasible, AlleleID® designs the minimum number of probes possible to detect all the sequences in an alignment. The program designs probes for the majority consensus first and then for the minority consensus and remaining individual sequences. This design is very useful, especially for studying phylogenetic relationships among sequences obtained from different geographical locations.
Taxa Specific/Cross Species Identification: Minimal Probes with Optimal Primers
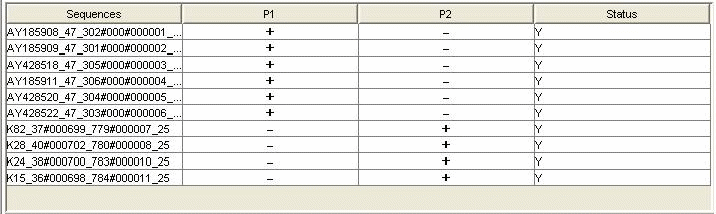
Y stands for the sequences that can be identified


